|
|||||||||||||||||
Broadly speaking we are interested in how the genes of organisms transmit their information to the cellular biosynthetic machinery. We are especially interested in how this is accomplished in the most complex of organisms—eukaryotes in general, but especially in mammals. RNA polymerase II is the enzyme charged with initiating the transfer of information from the DNA in the nucleus to the cell cytoplasm via RNA. Amazingly, RNA polymerase not only carries out the intricate task of precisely transcribing the DNA, but it also oversees the elaborate network of events involved in processing the RNA that it makes. Our focus is on the functional interactions of the transcriptional machinery with the machineries responsible for cleavage & polyadenylation and for splicing.
Cleavage & polyadenylation: The poly(A) signal is a sequence in the RNA that recruits a large collection of proteins which then cleave the RNA at the poly(A) site and add the signature poly(A) tail. From a purely biochemical perspective these are two very simple tasks. Yet the protein apparatus required to do this is large and complicated. The reason for this complexity is that this is the step that defines the final extent of the message, and it is involved in regulatory interactions with virtually every other step of mRNA production and export, even directing the termination of transcription itself. It is an important molecular checkpoint that determines whether, when, where and how to culminate the definition of each mRNA. We are using both in vivo and in vitro systems to study the assembly of the cleavage & polyadenylation apparatus, with the focus on its role as the final checkpoint prior to release of the RNA from the site of transcription.
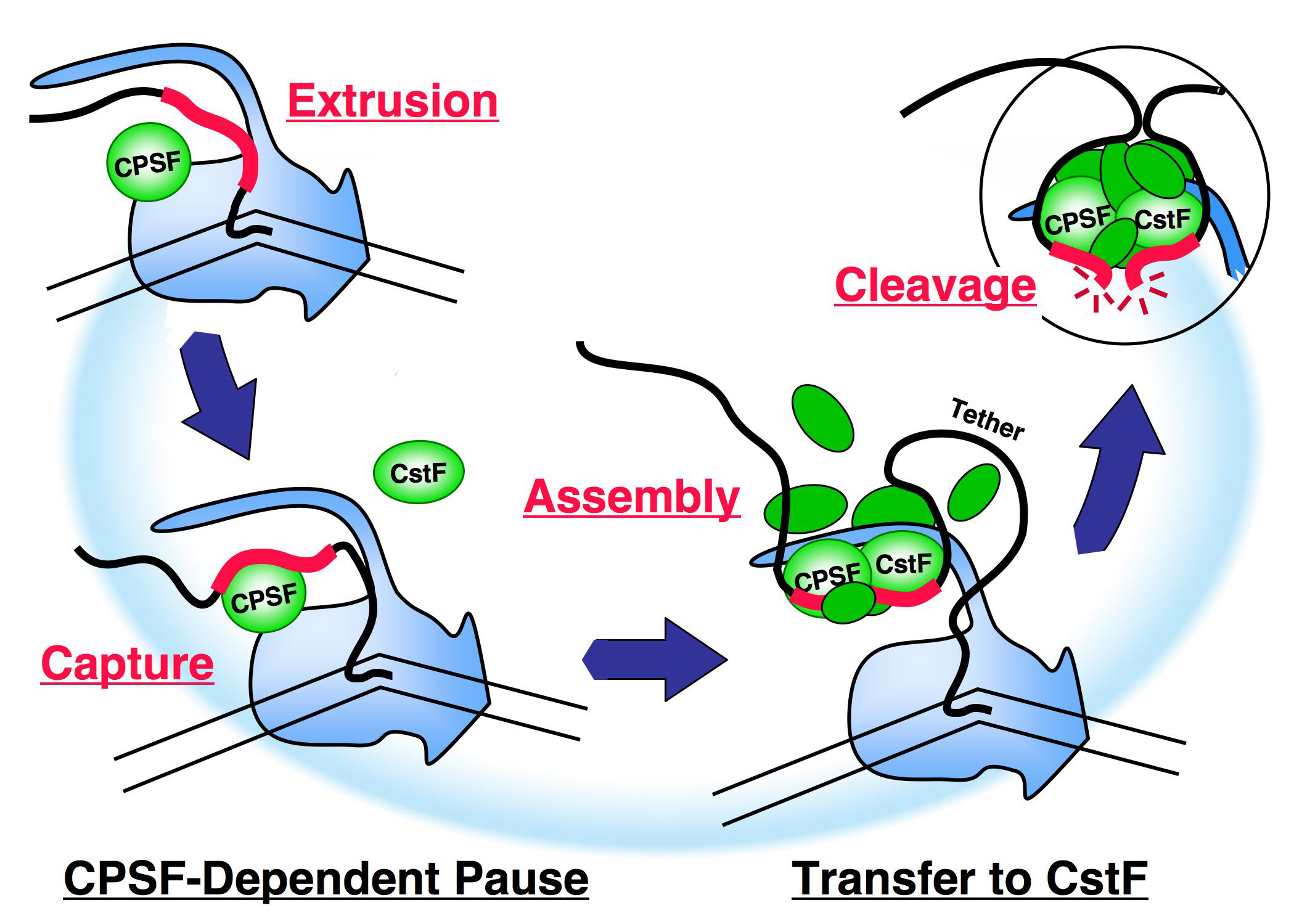
The cartoon on the right summarizes some of our recent results in terms of a speculative model. Cleavage/polyadenylation factor CPSF, riding the body of the polymerase, captures an emerging poly(A) signal and provokes the polymerase to pause. A second factor, CstF, is then also recruited and drives the formation of a CPSF-CstF complex which leads to a reorganization of the apparatus, probably on the CTD. Assembly continues, but these early stages of processing complex assembly are very tenuous and the assembling factors are easily lost. An important margin of stability is provided by the tether from the poly(A) signal to the polymerase. If the assembling apparatus becomes sufficiently mature to cling to the polymerase on its own before the tether becomes too long, cleavage of the RNA, termination of transcription and other downstream events will occur. The reorganization of the apparatus during assembly, and its precarious stability are important elements in its ability to coordinate regulatory input as a key checkpoint in mRNA maturation.
Splicing: It has become abundantly clear in recent years that alternative splicing is responsible for much of the diversity of the human proteome. Moreover, it is increasingly apparent that the specificity of alternative splicing is often rooted in the coupling of splicing to transcription. A major step forward in our ability to study coupling has been made possible by the development of an in vitro system capable of reproducing the functional interactions among transcription, splicing and cleavage/polyadenylation, in which all appear coupled to the other. We are using this system to explore the molecular basis for the role of transcription in alternative splicing and to study the fundamental mechanism of coupling itself.
Tsao, D.C. Park, N.J., Nag, A. & H.G. Martinson 2012. Prolonged α-amanitin treatment of cells for studying the polymerase CTD causes degradation of DSIF 160 and other proteins. RNA 18, 222-229. Martinson, H. G. 2011. An active role for splicing in 3′-end formation. Wiley Interdisciplinary Reviews: RNA, 2: 459–470. Kazerouninia, A., Ngo, B. & H.G. Martinson 2010. Poly(A) signal-dependent degradation of unprocessed nascent transcripts accompanies poly(A) signal-dependent transcriptional pausing in vitro. RNA, 16: 197-210. Rigo, F. and H.G. Martinson 2009. Polyadenylation releases mRNA from RNA polymerase II in a process that is licensed by splicing. RNA, 15: 823-836. Rigo, F. and H.G. Martinson 2008. Functional Coupling of Last-Intron Splicing and 3'-End Processing to Transcription In Vitro: the Poly(A) Signal Couples to Splicing before Committing to Cleavage. Mol. Cell. Biol.,
28:849-862. Nag, A., Narsinh, K., Kazerouninia, A. and H. G. Martinson 2006. The conserved AAUAAA hexamer of the poly(A) signal can act alone to trigger a stable decrease in RNA polymerase II transcription velocity. RNA, 12:1534-1544. Rigo, F., Kazerouninia, A., Nag, A. and H.G. Martinson 2005. The RNA Tether from the Poly(A) Signal to the Polymerase Mediates Coupling of Transcription to Cleavage and Polyadenylation. Molecular Cell, 20:733-745. Park, N.J., Tsao, D.C., and H.G. Martinson 2004. The Two Steps of Poly(A)-Dependent Termination, Pausing and Release, Can Be Uncoupled by Truncation of the RNA Polymerase II CTD. Mol. Cell. Biol., 24:4092-4103. Kim, S.J. and H.G. Martinson 2003. Poly(A)-Dependent Transcription Termination: Continued Communication of the Poly(A) Signal With the Polymerase Is Required Long After Extrusion In Vivo. J. Biol. Chem. 278:41691-41701. Tran, D.P., S.J. Kim, N.J. Park, T.M. Jew, and H.G. Martinson 2001. Mechanism of poly(A) signal transduction to RNA polymerase II in vitro. Mol. Cell. Biol. 21:7495-7508. Yeung, G., L.M. Choi, L.C. Chao, N.J. Park, D. Liu, A. Jamil, and H.G. Martinson 1998. Poly(A)-driven and poly(A)-assisted termination: two different modes of poly(A)-dependent transcription termination. Mol. Cell. Biol. 18:276-89.
Department of Chemistry & Biochemistry
:Last Update: 4/16/12 |
|||||||||||||||||