Tools
Resources
Open access research resources created by the Torres lab with generous funding
from the National Science Foundation, NSF MCB1243645.
CANVS: CANVS Download
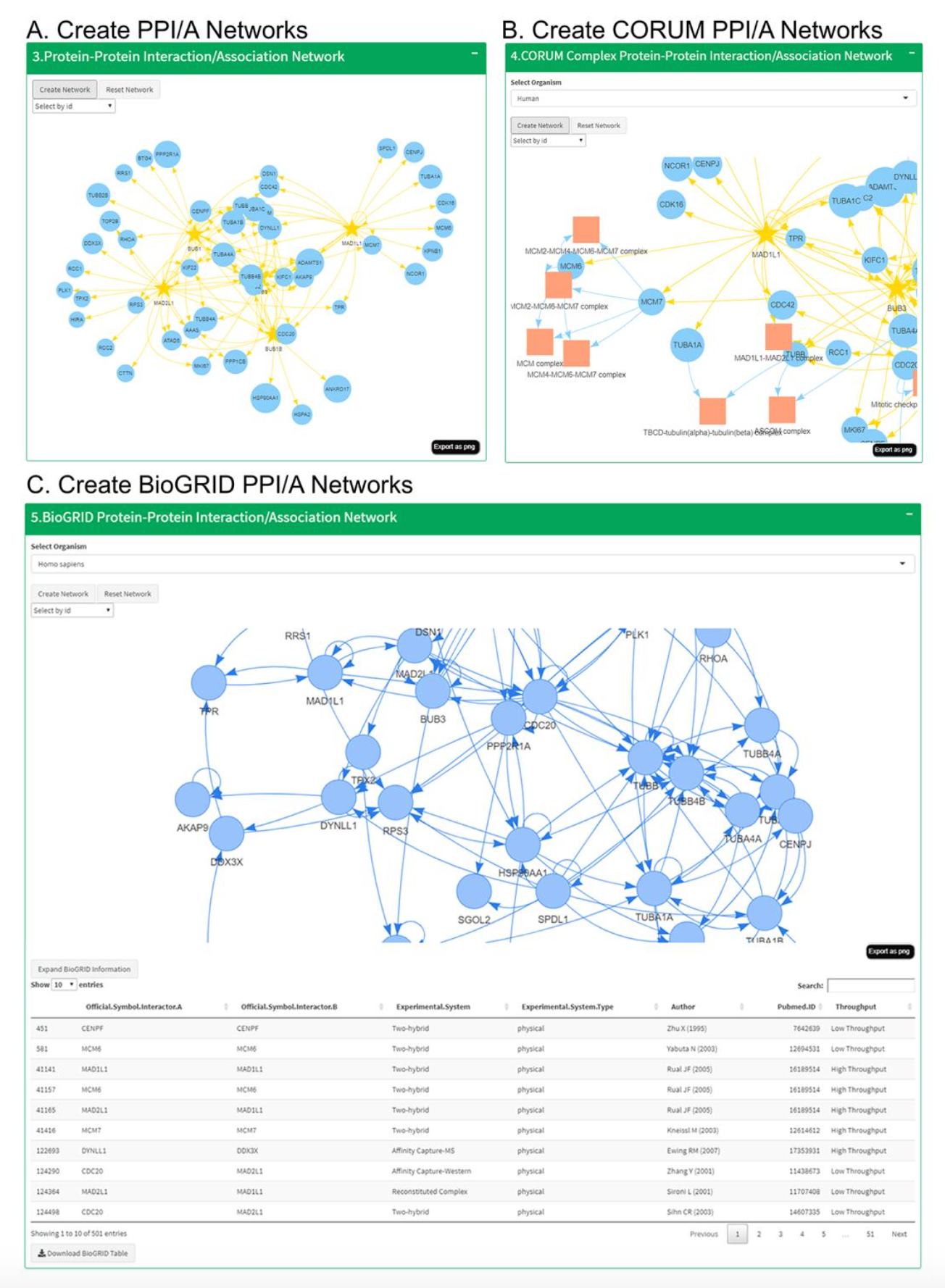
CANVS is a computational pipeline that cleans, analyzes, and visualizes mass
spectrometry-based interactome/association data. CANVS is wrapped as an
interactive Shiny dashboard, allowing users to easily interface with
the pipeline. With simple requirements, users can analyze complex experimental
data and create PPI/A networks. The application integrates systems biology
databases like BioGRID and CORUM to contextualize the results. Furthermore,
CANVS features a Gene Ontology tool that allows users to identify relevant
GO terms in their results and create visual networks with proteins associated
with relevant GO terms. Overall, CANVS is an easy-to-use application that benefits
all researchers, especially those who lack an established bioinformatic pipeline
and are interested in studying interactome/association data.
CSNAP web server: CSNAP
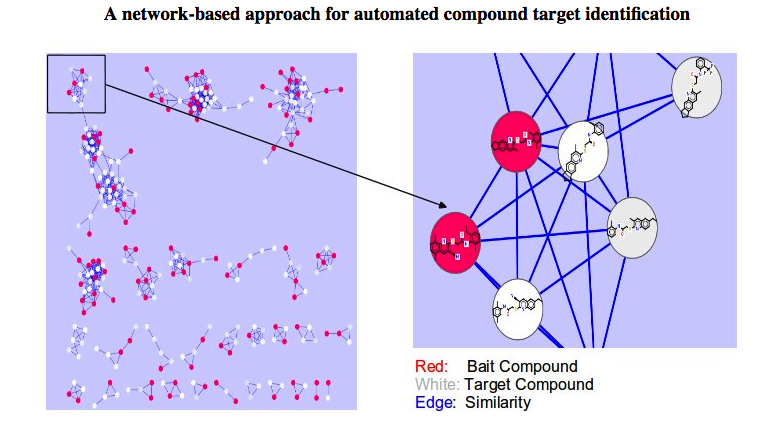
CSNAP (Chemical Similarity Network Analysis Pull-down) is a computational approach
for compound target identification based on network chemical similarity graphs.
Query and reference compounds are populated on the network connectivity map and
a graph-based neighbor counting method is applied to rank the consensus targets
among the neighborhood of each query ligand. The CSNAP approach can facilitate
high-throughput target discovery and off-target prediction for any compound set
identified from phenotype-based or cell-based chemical screens.
Generation of Inducible LAP-tagged stable cell lines and LAP biochemical purifications video:

Video describe a method for generating localization and affinity purification (LAP)-tagged inducible stable cell lines for investigating protein function, spatiotemporal subcellular localization and protein-protein interaction networks.
Published article:
Bradley M., Ramirez I., Cheung K., Gholkar A.A., and Torres J.Z. “Inducible LAP-tagged Stable Cell Lines for Investigating Protein Function, Spatiotemporal Localization and Protein Interaction Networks”, Journal of Visualized Experiments 2016 Dec. 24, (118):1-9, e54870, doi:10.3791/54870. PMID:28060263
Torres Lab Home | UCLA | Department of Chemistry & Biochemistry | Biochemistry Molecular Structural Biology (BMSB)