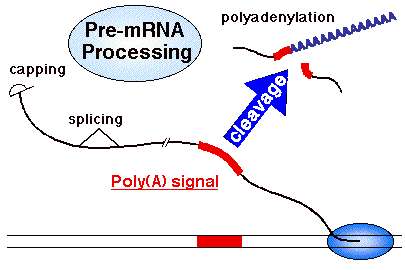
Capping, splicing and 3'-end formation all
impinge on the process of transcription termination by RNA polymerase II. Our attention is focussed on 3'-end formation because the effects of the upstream
processes on transcription appear to be chanelled through this event.
In higher eukaryotes mRNA 3'-end formation occurs in two
ways. For the
replication-dependent histone mRNAs 3'-end formation occurs by a
cleave-and-trim process. For all
other mRNAs 3'-ends are formed by cleavage and polyadenylation as illustrated
here.
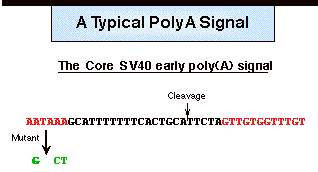 Cleavage and polyadenylation is directed
by a poly(A) signal in the RNA. The
core poly(A) signal for vertebrate pre-mRNAs consists of two recognition elements
(red) flanking a cleavage-polyadenylation site.
Typically, an almost invariant AAUAAA hexamer lies 20-50 nucleotides
upstream of a more variable element rich in U or GU residues.
Cleavage of the nascent transcript occurs between these two elements
and is coupled to the addition of up to 250 adenosines to the 5' cleavage
product.
Usually mutation (e.g. in green) of the hexamer completely inactivates the
poly(A) signal.
Cleavage and polyadenylation is directed
by a poly(A) signal in the RNA. The
core poly(A) signal for vertebrate pre-mRNAs consists of two recognition elements
(red) flanking a cleavage-polyadenylation site.
Typically, an almost invariant AAUAAA hexamer lies 20-50 nucleotides
upstream of a more variable element rich in U or GU residues.
Cleavage of the nascent transcript occurs between these two elements
and is coupled to the addition of up to 250 adenosines to the 5' cleavage
product.
Usually mutation (e.g. in green) of the hexamer completely inactivates the
poly(A) signal.
In vitro, cleavage is mediated by a protein complex that can
be separated into five factors. Two
of these factors are the cleavage and polyadenylation specificity factor (CPSF)
which binds the AAUAAA motif and the cleavage stimulation factor (CstF) which
binds the downstream U-rich element. In vivo, additional factors are required because of the coupling
to transcription.
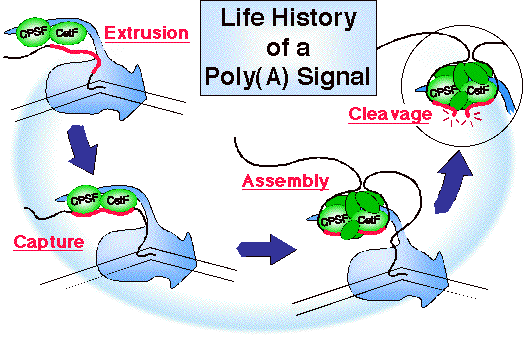 As the poly(A) signal is extruded from the polymerase it is captured by a subset of (green) cleavage and polyadenylation factors that
have already been recruited to the polymerase CTD. Then
assembly continues as more factors join the complex.
Ultimately cleavage occurs (illustrated here), followed by polyadenylation
(not shown).
As the poly(A) signal is extruded from the polymerase it is captured by a subset of (green) cleavage and polyadenylation factors that
have already been recruited to the polymerase CTD. Then
assembly continues as more factors join the complex.
Ultimately cleavage occurs (illustrated here), followed by polyadenylation
(not shown).